Diversity of Xanthomonas from brassicas in Nepal
By Steve Roberts, 10 Feb 2010
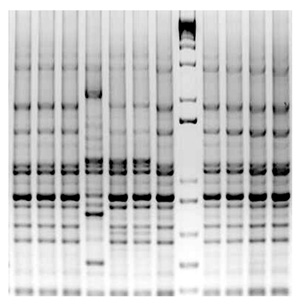
ABSTRACT Jensen, B. D., Vicente, J. G., Manandhar, H. K., and Roberts, S. J. 2010. Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable Brassica fields in Nepal. Plant Dis. 94:298-305. Black rot caused by Xanthomonas campestris pv. campestris was found in 28 sampled cabbage fields in five major cabbage-growing districts in Nepal in 2001 and in four cauliflower fields in two districts and a leaf mustard seed bed in 2003. Pathogenic X. campestris pv. campestris strains were obtained from 39 cabbage plants, 4 cauliflower plants, and 1 leaf mustard plant with typical lesions. Repetitive DNA polymerase chain reaction-based fingerprinting (rep-PCR) using repetitive extragenic palindromic, enterobacterial repetitive intergenic consensus, and BOX primers was used to assess the genetic diversity. Strains were also race typed using a differential series of Brassica spp. Cabbage strains belonged to five races (races 1, 4, 5, 6, and 7), with races 4, 1, and 6 the most common. All cauliflower strains were race 4 and the leaf mustard strain was race 6. A dendrogram derived from the combined rep-PCR profiles showed that the Nepalese X. campestris pv. campestris strains clustered separately from other Xanthomonas spp. and pathovars. Race 1 strains clustered together and strains of races 4, 5, and 6 were each split into at least two clusters. The presence of different races and the genetic variability of the pathogen should be considered when resistant cultivars are bred and introduced into regions in Nepal to control black rot of brassicas.
Tags: black rot; brassicas; Xanthomonas; disease; Nepal; fingerprinting